Multi-modal ML for BBD
Stratifying risk for breast cancer using genomic data from benign breast biopsies
The Big Picture
Breast cancer is one of the most common cancers worldwide, and benign breast disease (BBD) is a known risk factor. But here’s the twist: only 5% of BBD cases are the high-risk atypical hyperplasia type. What about the other 95%? Can we dig into the DNA of these “low-risk” cases and uncover patterns that predict breast cancer?
What We Did
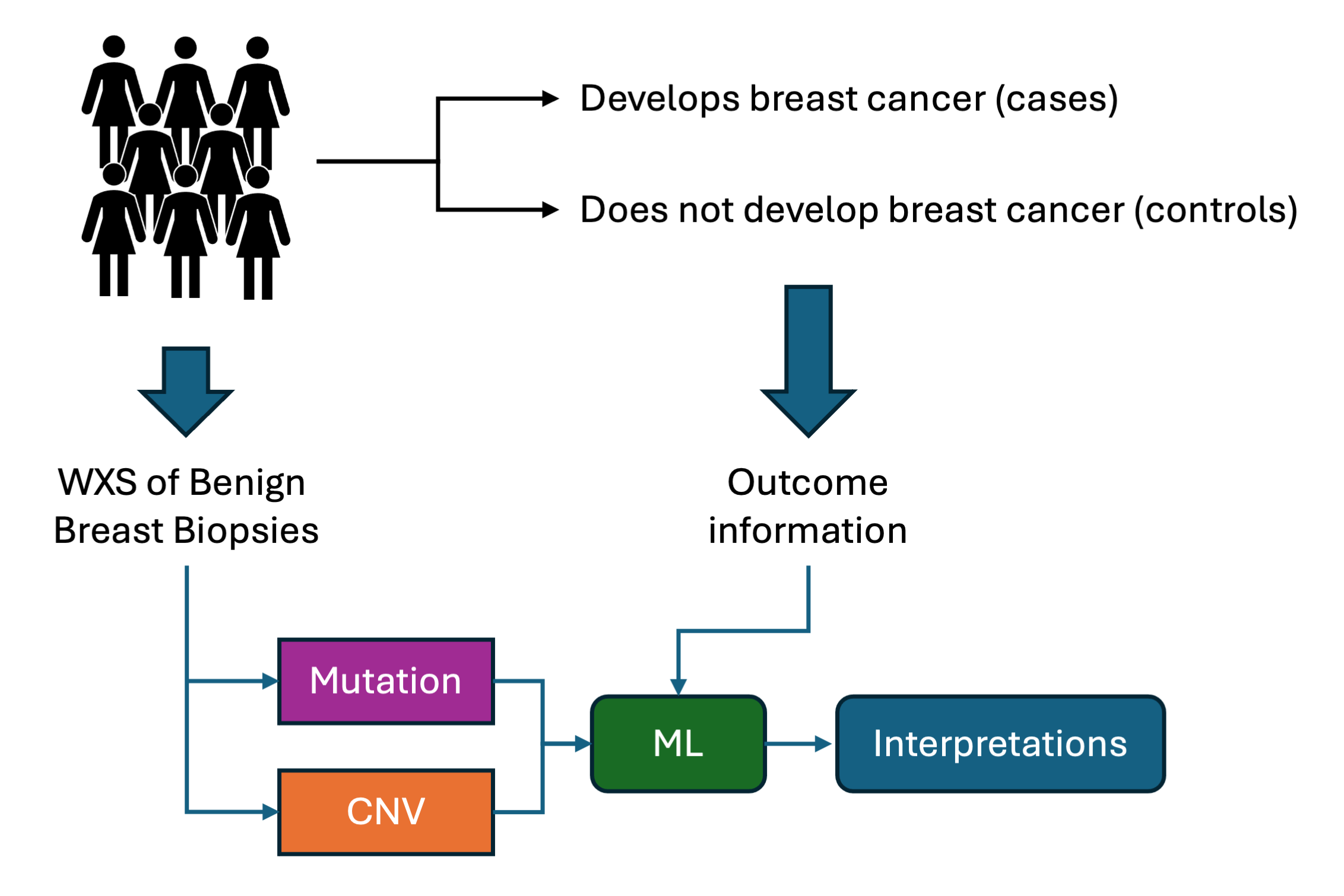
We analyzed whole-genome sequencing (WGS) data from 240 BBD samples, alongside a subset of 91 matched saliva samples. Our approach included:
- Decoding Mutational Patterns: Extracting mutational signatures like single-base substitutions (SBS), double-base substitutions (DBS), and insertion-deletions (ID).
- Identifying Primising DNA Copy-Number Variations (CNVs): Identifying regions with recurrent CNVs in our cohort.
- Developing Risk Models: Combining mutations and CNVs in a multi-modal machine learning (ML) approach to predict breast cancer risk. We tested three different fusion approaches for our two modalities.
What We Found
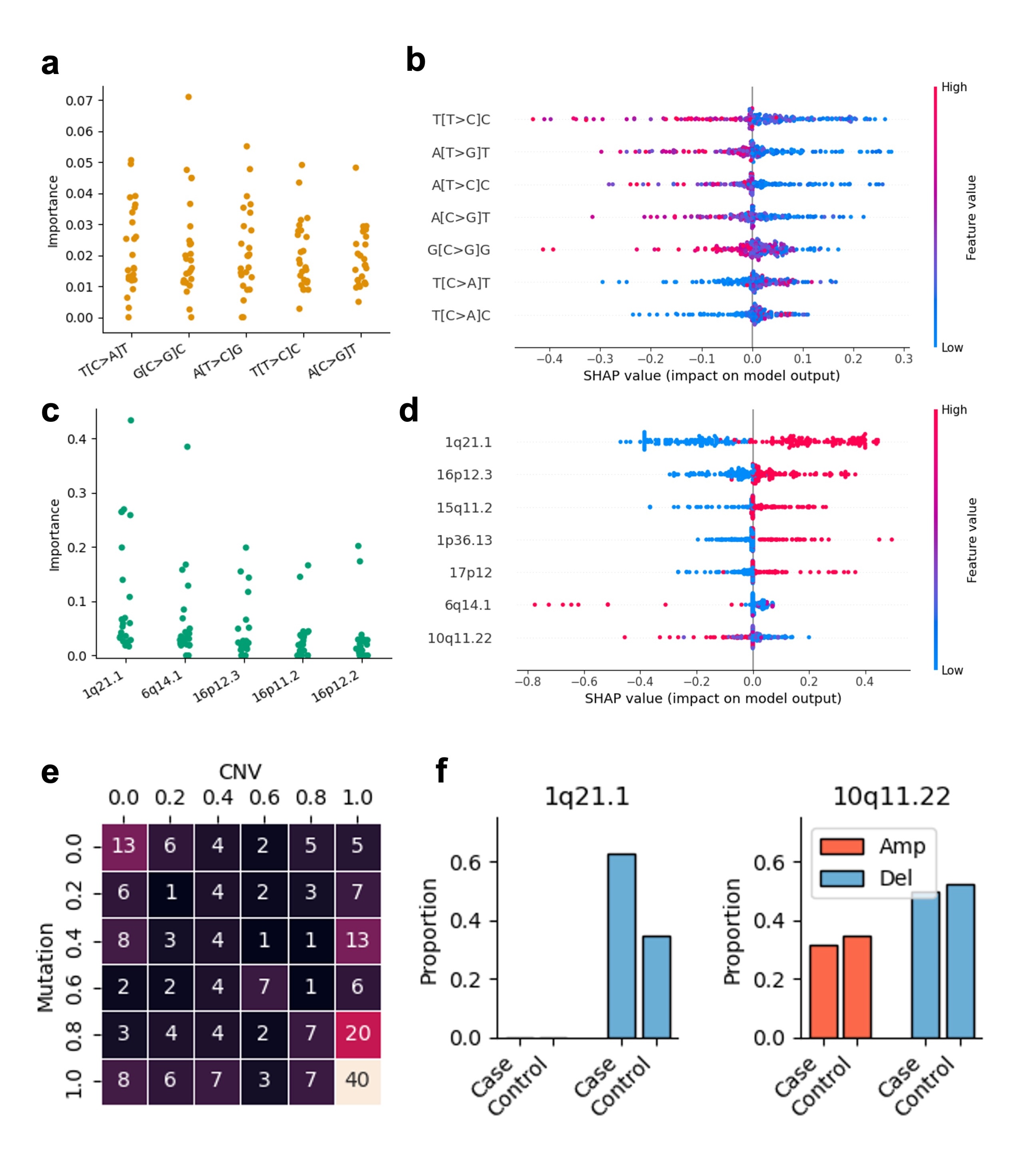
- Mutation Signatures: We identified 9 SBS, 1 DBS, and 3 ID signatures, some linked to DNA mismatch repair issues—a hallmark of cancer risk.
- Copy Number Variations: Found hundreds of recurrent DNA changes, including a BRCA1 deletion that was specifically recurrent in the high-risk population.
- Model Performance: Our models achieved a decent predictive accuracy (ROC-AUC ~0.60). But here’s the catch: 13 samples were consistently misclassified on all the folds. These samples did not show statistically significant correlations with patient age, library size, or biopsy year, suggesting some unknown sample variability that warrants further investigation.
Where Do We Go From Here?
Our findings suggest that our current approach is likely limited in its clinical utility due to its modest performance. What’s next?
- Adding epigenetics to the mix (Refer to my breast epigenetics project page for why I think this might be a good idea!)
- Exploring new ways to engineer features and/or fuse the two modalities of data –The genomic space is vast and noisy! It’s all about finding the right feature representations.
- Digging deeper into the samples that break the mold—are they simply outliers, or do they hold useful information?